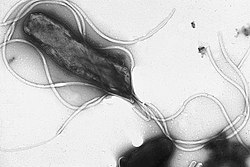
Flagellum
| We're all Homo here Evolution |
| Relevant Hominids |
| A Gradual Science |
| Plain Monkey Business |
The flagellum (plural: flagella) is a wiggly/spinny structure used by many single-celled organisms to move through a liquid medium. In eukaryotes, the more proper term is cilium (plural: cilia). The primary component of both prokaryotic (bacterial and archaeal) flagella is the protein flagellin.
There are three distinct groups of flagella, each of which has its own evolutionary pathway (convergent evolution). The bacterial flagellum is a helical filament that rotates like a screw. The archaeal flagellum is similar (but non-homologous) to the bacterial flagellum), and the eukaryotic flagellum/cilium is a whip-like structure that lashes back and forth.
Intelligent design proponents and creationists argue that the flagellum is so complex that it cannot have evolved on its own — necessitating the intervention of an intelligent designer.
And most importantly, it has a funny name. Come on, say it. You know you want to.
Explanation of terms[edit]
There are a large number of systems that single-celled organisms use to move or swim. Those systems that work by sticking something large out of the cell and moving it are often called "flagella." The word flagellum was originally the Latin word for whip.
Although the same term is used, there are three (known) kinds of "flagella" that are very different in detail. They are often confused because terminology is often used inconsistently.
- Bacteria have flagella. The motor is at the base and they rotate. They are always called "flagella" or "bacterial flagella".
- Archaea (also known as archaebacteria) have flagella. The motor is at the base and they rotate. But, despite early assumptions of relatedness to bacterial flagella based on these similarities, they are very different in detail. They are always called "flagella" or "archaeal flagella".
- Eukaryotes have a tubulin-based organelle that does not rotate. Instead, this organelle bends all along its length, powered by hundreds of dynein motor proteins. This organelle is called variously a flagellum, cilium, or undulipodium, or sometimes other names. This leads to a great deal of confusion for people new to the topic. Usage typically works like this:
- Most microbiologists call it a "flagellum" if the eukaryotic cell has one or a few long appendages (such as sperm cells), and call it a "cilium" if the cell has many shorter appendages (such as a paramecium)
- Some people point out that eukaryotic flagella and cilia have fundamentally the same 9+2 tubulin structure (usually), dyneins, etc., and are really essentially the same thing. They propose that the word "cilium" be used for both kinds of structures, and that "flagellum" be reserved for the prokaryote organelles. This is the position and usage of Thomas Cavalier-Smith, and this usage is followed by Michael Behe in Darwin's Black Box
- Lynn Margulis and her followers (relatively few but published widely) call the eukaryotic structure a "undulipodium", both in order to distinguish it from the prokaryote organelles and to emphasize their symbiotic theory for the origin of the organelle.
Importance to intelligent design[edit]
As with many aspects of science that creationists do not understand, the evolution of the flagellum is always on their "questions science cannot answer"[1][2] lists, specifically with regards to irreducible complexity as Michael Behe postulates it: How could the flagellum have appeared? There is nothing smaller than a fully functioning flagellum, they would state. "Removing any of its parts makes it non-functional."
Biochemist Michael Behe, of Lehigh University and Senior Fellow of the Discovery Institute, wrote a 1996 book entitled Darwin's Black Box, in which claimed that "irreducibly complex" (IC) systems, systems which require several parts to function, were either impossible (or very unlikely) to reach via natural evolutionary mechanisms, and therefore must have been designed by an intelligence.
Behe's first two major example systems were the eukaryotic flagellum/cilium (henceforth cilium) and the bacterial flagellum. Behe asserted that scientists had no idea how such structures evolved. This argument has been wildly popular among anti-evolutionists, who in particular have latched onto the bacterial flagellum as an icon of ID.
The flagellum became so famous a "body part", that it has appeared in law court, specifically Kitzmiller v. Dover, when Behe introduced the idea that tides go in, tides go out, you can't explain that "The flagellum is so precisely constructed, no single part can be removed without the whole thing failing. Evolution cannot explain how that happens."
In The Flagellum Unspun Ken Miller recreates the argument he used against Behe in the Dover Courthouse.[3]
Despite the fact that biologist Ken Miller and others have specifically shown exactly how a flagellum could have evolved, as distinct, critical, functional and useful steps, creationists continue to use the flagellum as one of their oft-repeated "self-evident" arguments. A video of Miller's arguments on the flagellum can be seen here.
Eukaryotic[edit]
Description[edit]
The eukaryotic flagellum is completely different from the prokaryote flagella in structure and in evolutionary origin. The only thing that the bacterial, archaeal, and eukaryotic flagella have in common is that they stick outside of the cell and wiggle to produce propulsion.
A eukaryotic flagellum is a bundle of nine fused pairs of microtubules called "doublets" surrounding two central single microtubules (the so-called 9+2 structure; also called the "axoneme"). At the base of a eukaryotic flagellum is a microtubule organizing center about 500 nanometers long, called the basal body or kinetosome. The flagellum is encased within the cell's plasma membrane, so that the interior of the flagellum is accessible to the cell's cytoplasm. This is necessary because the flagellum's flexing is driven by the protein dynein bridging the microtubules all along its length and forcing them to slide relative to each other, and ATP must be transported to them for them to function.
Evolution[edit]
There are two competing groups of models for the origin of the cilium:[4]
Symbiotic/endosymbiotic/exogenous models[edit]
These argue some version of the idea that the cilium evolved from a symbiotic spirochete that attached to a primitive eukaryote or archaebacterium. The modern version of the hypothesis was first proposed by Lynn Margulis as Sagan (1967) (Margulis was the first wife of the late Carl Sagan). The hypothesis, though very well publicized, was never widely accepted by the experts, in contrast to Margulis' successful arguments for the symbiotic origin of mitochondria and chloroplasts.
The only real point in favor of the symbiotic hypothesis is that there apparently actually are eukaryotes that use symbiotic spirochetes as their motility organelles (only inside termite guts, though, as far as I know, i.e., Trichonympha). While this is a flabbergasting example of co-option and the creativity and flexibility of biological systems, none of the proposed homologies that have been reported between cilia and spirochetes (e.g. Bermudes et al. 1987; Barth et al. 1991) have stood up to further scrutiny (e.g. Bermudes et al. 1994, Munson et al. 1993). The homology of tubulin to the bacterial replication/cytoskeletal protein FtsZ (see "Some web references on FtsZ-tubulin", below) would seem to clinch the case against Margulis, as FtsZ is apparently found native in archaebacteria (e.g. see Faguy and Doolittle, 1998), providing an endogenous ancestor to tubulin (as opposed to Margulis' hypothesis, that an archaebacterium acquired tubulin from a symbiotic spirochete — see Margulis et al., 2000 for the latest version).
At present the symbiotic hypothesis for the origin of cilia seems to be basically a pet idea of Margulis and a few of her associates. Margulis is, though, still strongly promoting and publishing a revised version of her hypothesis (e.g. the articles listed below, including a few freely available online in PNAS. Margulis' 1998 book Symbiotic planet: A New Look at Evolution has some frank autobiographical comments about her stubborn support of the symbiotic hypothesis for the origin of the cilium.
Some references on Margulis' endosymbiotic hypothesis[edit]
- Barth, A. L., Stricker, J. A. and Margulis, L. (1991). "Search for Eukaryotic Motility Proteins in Spirochetes — Immunological Detection of a Tektin-Like Protein in Spirochaeta-Halophila." BioSystems, V24(N4): 313-319 [2]
- Bermudes, D., Fracek, S. P. Jr., Laursen, R. A., Margulis, L., Obar, R. and Tzertzinis, G. (1987). "Tubulinlike protein from Spirochaeta bajacaliforniensis," in Annals of the New York Academy of Sciences: Endocytobiology III. New York, The New York Academy of Sciences. 503: 515-527 [3]
- Bermudes, D., Hinkle, G. and Margulis, L. (1994). "Do Prokaryotes Contain Microtubules." Microbiological Reviews, V58(N3): 387-400 [4] [Note 2]
- Bermudes, D., Margulis, L. and Tzertzinis, G. (1987). "Prokaryotic Origin of Undulipodia: Application of the Panda Principle To the Centriole Enigma," in Annals of the New York Academy of Sciences: Endocytobiology III. New York, The New York Academy of Sciences. 503: 187-197 [5]
- Chapman, M. J., Dolan, M. F. and Margulis, L. (2000). "Centrioles and kinetosomes: Form, function, and evolution." Quarterly Review of Biology, V75(N4): 409-429 [6]
- Corliss, J. O. (1980). "Objection to "undulipodium" as an inappropriate and unnecessary term." BioSystems, 12: 109-110 [7]
- Guerrero, R., Pedros-Alio, C., Esteve, I., Mas, J., Chase, D. and Margulis, L. (1986). "Predatory prokaryotes: predation and primary consumption evolved in bacteria." Proceedings of the National Academy of Sciences, 83: 2138-2142. [8]
- Hülsmann, N. (1992). "Undulipodium: End of A Useless Discussion." European Journal of Protistology, 28(3): 253-257. [Note 3]
- Margulis, L. (1992). "Protoctists and Polyphyly — Comment." BioSystems, V28(N1-3): 107-108. [9]
- Margulis, L. (1996). "Archaeal-Eubacterial Mergers in the Origin of Eukarya - Phylogenetic Classification of Life." Proceedings of the National Academy of Sciences of the United States of America, V93(N3): 1071-1076 [10]
- Margulis, L., Chase, D. and To, L. P (1979). "Possible evolutionary significance of spirochaetes." Proc R Soc Lond B Biol Sci, 204(1155): 189-198 [11]
- Margulis, Lynn (1980). "Undulipodia, flagella, and cilia." BioSystems, 12: 105-108 [12]
- Margulis, Lynn (1993). Symbiosis in cell evolution : microbial communities in the Archean and Proterozoic eons. New York, Freeman, pp. xxvii, 452. Link: [13]
- Margulis, Lynn (1998). Symbiotic planet : a new look at evolution. New York, Basic Books, pp. vi, 147. [14]
- Margulis, Lynn, Dolan, Michael F. and Guerrero, Ricardo (2000). "The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists." Proceedings of the National Academy of Sciences, 97(13): 6954-6959. [15] [Note 4]
- Maynard Smith, John and Szathmáry, Eörs (1995). The major transitions in evolution. Oxford, New York, W.H. Freeman Spektrum, pp. xiv, 346. [16]
- Munson, D., Obar, R., Tzertzinis, G. and Margulis, L. (1993). "The Tubulin-Like S1 Protein of Spirochaeta Is a Member of the Hsp65 Stress Protein Family." BioSystems, V31(N2-3): 161-167 [17]
- Sagan, Lynn (1967). "On the Origin of Mitosing Cells." Journal of Theoretical Biology, 14: 225-274. [Note 1]
- Szathmary, E. (1987). "Early evolution of microtubules and undulipodia." BioSystems, 20(2): 115-132 [18]
- Wheatley, D. N. (1982). The centriole, a central enigma of cell biology. Amsterdam, Elsevier Biomedical Press, pp. 1-232. [19]
- Note 1: Behe referenced this paper in Darwin's Black Box.
- Note 2: Behe cites this on p. 279 as a 1986 paper. The volume was published in 1987 and is so listed in databases, but the meeting was in 1986, which is probably what Behe was thinking of.
- Note 3: It is worth pointing out that this 1992 exchange between Cavalier-Smith and Margulis, described by Behe thusly:
"Margulis and Cavalier-Smith have clashed in print in recent years6 [cites the 1992 papers]. Each points out the enormous problems with the other's model, and each is correct." (DBB, p. 69)
- had essentially nothing to do with the origin of the cilium. Cavalier-Smith's 15-page article was essentially entirely about protozoan taxonomy and how often well-accepted symbioses, like those of mitochondria and chloroplasts, had occurred (and he did criticize Margulis, and others, along these lines). Cavalier-Smith devotes only two sentences to commenting on Margulis' spirochete hypothesis, and these are merely to say that he won't be discussing it because he regards it an "implausible speculation". Margulis' brief reply (one and a half pages) is also mostly about taxonomy and symbiotic questions. She takes a few sentences to cite some of the evidence she thought supported the spirochete hypothesis (it was disconfirmed in the literature in following years), but she spends no time criticizing Cavalier-Smith's scenario. Critiques (in my opinion marginal) of Cavalier-Smith's earlier (1978 and 1982) papers can be found in Szathmary (1987) and Bermudes et al. (1987), but that same year Cavalier-Smith (1987b) revised his earlier model and integrated it with his larger work on the origin of eukaryotes.
- I have not found any critiques of Cavalier-Smith's (1987b) model, but in his 1992 (b) paper "Origin of the Cytoskeleton" Cavalier-Smith refers to his earlier work as the "classical" autogenous hypothesis. This (1992b) paper, not cited by Behe, is where one will find the actual detailed criticisms of Margulis' and Szathmary's models by Cavalier-Smith, which are in my opinion devastating, although documenting all of this will require quite a long review article.
- Note 4: This book discusses the origin and evolution of numerous key systems in biology, from the origin of life, to eukaryote mitosis, meiosis, and sex, to multicellularity and sociality. It also includes a good discussion of the centriole and the possibilities for its replication and origin (which is closely tied to the same questions for the cilium), but the authors (early spirochete-hypothesis supporters, both) do little discussion and no defense of the spirochete hypothesis, and although they do cite Szathmary (1987), one gets the impression that they do not remain strong supporters.
- More importantly for Behe, the book even discusses the challenge of irreducible complexity on the very first page (p. 3, now displayed online at amazon.com), although the actual term is not used. This is yet another book that Behe should have found, especially since he mentions other work by both authors in DBB, and the book was preceded by numerous scientific articles by the authors, and particularly since Behe makes extravagant, confident claims to the public about what books and articles do and do not exist.
Endogenous/Autogenous/direct filiation models[edit]
The cilium developed from pre-existing components of the eukaryotic cytoskeleton (which has tubulin, dynein, and nexin, used for other functions of course) (McQuade, 1977; Cavalier-Smith 1975, 1978, 1982) as an extension of the mitotic spindle apparatus (Cavalier-Smith, 1987b). The connection can still be seen, first in the various early-branching single-celled eukaryotes that have a microtubule basal body, where microtubules on one end form a spindle-like cone around the nucleus, while microtubules on the other end point away from the cell and form the cilium. A further connection is that the centriole, involved (somehow, scientists are unsure of the purpose of the centriole) in the formation of the mitotic spindle in many (but not all) eukaryotes, is homologous to the cilium, and in many cases is the basal body from which the cilium grows. As Cavalier-Smith noted in his papers on this topic (there was more than one, contra Behe), an obvious intermediate stage between spindle and cilium would be a non-swimming appendage made of microtubules with a selectable function like increasing surface area, helping the protozoan to remain suspended in water, and/or increasing the chances of bumping into bacteria to eat (or serving as a stalk attaching the cell to a solid substrate, the suggestion of Rizzotti, 1995, who otherwise advocates an endogenous scenario similar to Cavalier-Smith).
And one can't argue that such a non-swimming appendage is merely convenient imagination or unlikely to be selectable, as modern protists with analogous non-swimming microtubular appendages do exist and find them perfectly useful, the axopodia of phylum Actinopoda (see also [http://megasun.bch.umontreal.ca/protists/raphp/summary.html this page] on genus Raphidiophrys) being an oft-cited example. In his 1997 dissection of Behe ("The blind biochemist" in Trends in Ecology and Evolution), Cavalier-Smith noted, "[Behe] does not mention the evidence that...other motility organelles much simpler than cilia, for example, protozoan axostyles, evolved from bundles of microtubules by acquiring the capacity to bend, which he implies is impossible."
Regarding the origin of the individual protein components, an interesting paper on the evolution of dyneins (Gibbons, 1995; see also Asai and Koonce, 2001) shows that the more complex protein family of cilial dynein has an obvious ancestor in a simpler cytoplasmic dynein (which itself appears to be a result of a four-fold duplication of a smaller motif). Below I list several papers] on the origin of the cytoskeleton, if you're wondering where that came from. And recently, long-standing suspicions that tubulin was homologous to FtsZ (based on very weak sequence similarity and some behavioral similarities), mentioned by Cavalier-Smith in his 1997 review of Behe, were impressively confirmed in 1998 by the independent resolution of the 3-dimensional structures of the two proteins (several detailed webpages are listed below).
- [Note 1] refers to papers that Behe referenced. The other notes are the ones he missed, or that were published 1996 or later.
Some references to the work of Cavalier-Smith[edit]
(...and others on the origin of the cilium, cytoskeleton, and components thereof (and a few on related aspects of Cavalier-Smith's work on the origin of the cell)
- Asai, D. J. and Koonce, M. F. (2001). "The dynein heavy chain: structure, mechanics and evolution." Trends in Cell Biology, V11(N5): 196-202. [20]
- Cavalier-Smith, T. (1975). "The origin of nuclei and of eukaryotic cells." Nature, 256: 463-468.
- [Note 1] Cavalier-Smith, T. (1978). "The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella." Biosystems, 10(1-2): 93-114. i[21]
- Cavalier-Smith, T. (1982). "The evolutionary origin and phylogeny of eukaryote flagella." Symposia of the Societyfor Experimental Biology, 35(5896): 465-493. [22]
- Cavalier-Smith, T. (1986). "Cilia Versus Undulipodia." Bioscience, 36(5): 293-293.
- [Note 3] Cavalier-Smith, T. (1987a). "The Origin of Cells: A Symbiosis between Genes, Catalysts, and Membranes." Cold Spring Harbor Symposia on Quantitative Biology: The Evolution of CatalyticFunction, LII: 805-824. [23]
- Cavalier-Smith, T. (1987b). "The origin of eukaryotic and archaebacterial cells." Annals of the New York Academyof Sciences: Endocytobiology III, 503(6111): 17-54. [24]
- [Note 2] Cavalier-Smith, T. (1987c). "The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies." Annals of the New York Academy of Sciences: Endocytobiology III, 503(6111):55-71. [25]
- Cavalier-Smith, T. (1988). "Origin of the Cell Nucleus." Bioessays, 9(2-3): 72-78. [26]
- Cavalier-Smith, T. (1991). "Archamoebae the Ancestral Eukaryotes?" Biosystems, 25(1-2): 25-38. [27]
- Cavalier-Smith, T. (1991). "The Evolution of Prokaryotic and Eukaryotic Cells," in Fundamentals of Medical CellBiology: Evolutionary Biology. E. Edward Bittar. London, JAI Press. 1:217-272.
- [Note 1] Cavalier-Smith, T. (1992). "The Number of Symbiotic Origins of Organelles." Biosystems, 28(1-3):91-106. [28]
- Cavalier-Smith, T. (1992b). "Origin of the cytoskeleton," in The Origin and evolution of the cell: Conferenceon the Origin and Evolution of Prokaryotic and Eukaryotic Cells. Shimoda, Japan April 22-25, 1992. H. Matsuno Hartman, K. Singapore, World ScientificPublishing Co.: 79-106. [29]
- Cavalier-Smith, T. (2001). "Obcells as proto-organisms: Membrane heredity, lithophosphorylation, and the originsof the genetic code, the first cells, and photosynthesis." Journal of Molecular Evolution, V53(N4-5): 555-595. [30]
- Doolittle, Russell F. (1995). "The Origins and Evolution of Eukaryotic Proteins." Philosophical Transactions:Biological Sciences (Evolution of Eukaryotic Cellular Processes), 349(1329):235-240. [31]
- Egelman, Edward H. (1998). "Tubulin Family: Kinship of key proteins across phylogenetic domains." CurrentBiology, 8: R288-R290. [32]
- Faguy, D. M. and Doolittle, W. F. (1998). "Cytoskeletal proteins: The evolution of cell division." Current Biology, V8(N10):R338-R341. [33]
- Gibbons, I. R. (1995). "Dynein family of motor proteins: Present status and future questions." Cell Motilityand the Cytoskeleton, 32(2): 136-144. [34]
- McQuade, A. B. (1977). "Origins of the Nucleate Organisms." Quarterly Review of Biology, 52(3):249-262. [35]
- Mitchison, T. J. (1995). "Evolution of a dynamic cytoskeleton." Philosophical Transactions: Biological Sciences(Evolution of Eukaryotic Cellular Processes), 349: 299-304. [36]
- Nasmyth, Kim (1995). "Evolution of the Cell Cycle." Philosophical Transactions: Biological Sciences (Evolutionof Eukaryotic Cellular Processes), 349(1329): 271-281. [37]
- Rizzotti, Martino (1995). "Cilium: Origin and 9-Fold Symmetry." Acta Biotheoretica, 43(3): 227-240. [38]
- van den Ent, F., Amos, L. A. and Lowe, J. (2001). "Prokaryotic origin of the actin cytoskeleton." Nature, V413(N6851): 39-44. [39]
- Note 1: Behe referenced this paper in Darwin's Black Box.
- Note 2: This 1987(c) article, "The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies," is mentioned by Behe in footnote 4 of chapter 5 (DBB, p. 155, footnote on p. 281). Chapter 5 of DBB is on vesicular transport and protein translocation, and Behe says he found this article along with several others after failing to find anything via computer search: "Slogging through the literature the old-fashioned way turns up a few scattered papers that speculate on how gated transport between compartments of a eukaryotic cell might have developed [footnote 4 is here]." (Behe 1996, 114-115) Regarding vesicular transport, I have little knowledge of the subject so will refrain from judgement (but see note 3, below).
- But returning to the cilium, the interesting point is that Behe -- if he looked up this Cavalier-Smith paper from pages 55-71 of volume 503 of Annals of the New York Academy of Sciences -- cannot have failed to notice Cavalier-Smith's other paper ("The origin of eukaryotic and archaebacterial cells", 1987b) in the same volume, on pages 17-54. And this paper updates Cavalier-Smith's 1978 and 1982 work on the evolutionary origin of the cilium, by placing the origin of the cilium in the context of the origin of eukaryote mitosis, explicitly deriving the cilium from the primitive mitotic spindle, which provides a base and nucleation center for the microtubules. This answers the "how did the microtubules get pointed perpendicular to the cell membrane" question Behe raises, as well as helps to explain the peculiar connections found in modern eukaryotes between the cilium, the centriole, and mitosis. Anyway, as I said this topic requires it's own large FAQ, but the point for now is that Behe should have mentioned Cavalier-Smith's work on the origin of the cilium after his 1978 paper.
- Here is Cavalier-Smith's (1997) opinion on Behe's treatment of the cilium subject:
[W]hen criticizing existing evolutionary explanations,
Behe uses intellectually dishonest double standards. He dismisses my first treatment of the origin of cilia2 [footnote 2: the 1978 paper] as non-quantitative and therefore 'utterly useless', and ignores my later work on the topic3,4 [these are the 1982 and 1987b papers, respectively]. But it does not worry him that his empty, religious notion of 'intelligent design' is equally non-quantitative; worse still, lacking in even qualitative detail of what did the designing, and how the hypothetical design was executed, it explains nothing. He states that 'if a theory claims to be able to explain some phenomenon but does not even generate an attempt at an explanation is should be banished' and 'without details, discussion is doomed to be unscientific and fruitless'. If he had applied these strictures to his panacea of 'intelligent design'
we would have been spared this worthless book.
- Note 3: This volume of the Cold Spring Harbor Symposia on Quantitative Biology is mentioned by Behe in DBB (p. 179). Of the articles in it, Behe says that about "two-thirds...are simply overviews of what was going on in the author's lab at the time, with little or no attempt to tie it into the theme of the book." Having looked at the volume, this alone is a highly debatable statement. But Behe continues: "Of the remaining papers, most are sequence analyses, and some are concerned with prebiotic chemistry or simple catalysts (not the complex machinery of known organisms)."
- But strangely Behe failed to mention the 20-page article by Cavalier-Smith (1987a) on the origin of cells. The article addresses (among other things) the origin of protein translocation. Here is Cavalier-Smith's (1997) take on this situation (Ref 3 is Cavalier-Smith 1987b; Ref 7 is Cavalier-Smith 1987a, Ref 6 is Blobel 1980):
Behe, ignorant of much of the literature, claims that no scientist has ever discussed the origin of vesicle targeting (actually discussed in Ref. 3 [Cavalier-Smith, 1987b], not cited by Behe, though the most detailed one on the origin of eukaryotic biochemical properties) or protein translocation (see Refs 6 [Blobel, 1980, PNAS, v.77, 1496-1500] and 7 [Cavalier-Smith, 1987a]), the most detailed discussion of the origin of the most basic complex cellular biochemical properties, which he deceitfully ignored despite citing the volume containing it as 'evidence' that no paper has ever been published on the subject!). Maybe he did not want his readers to find the papers (Refs 3 and 7) that most clearly show how one can explain (in outline at least -- obviously they are not the final answer) the origins of complex biochemical and cellular structures in logical steps using mutation, selection and detailed phylogenetic arguments. His ignorant assumption that the origin of a protoSRP would have killed the cell is refuted by the absence of the translation arrest domain in the eubacterial signal recognition partical (SRP) RNA8 [Poritz, 1989, Cell, 4-6], which provides a simpler ancestor to the more complex archaebacterial/eukaryotic particle. The problem he raises [p. 112] for the origin of secreted eukaryotic glycoproteins is spurious, because the sugar must have been added to the protein on the non-cytosolic side of the membrane in the common ancestor of eukaryotes and archaebacteria, even before the ER (endoplasmic reticulum) evolved, since it is added thus at the archaebacterial cell surface, as any good biochemist should have known, even without reading my discussion of the origin of the ER (Ref. 3).
- Notably, Behe also failed to mention two other papers in the very same volume as the Cavalier-Smith (1987a) paper in Cold Spring Harbor Symposia on Quantitative Biology. One is even by Russell Doolittle, on the evolution of vertebrate blood coagulation.
- Blake, C. C. F., Harlos, K. and Holland, S. K. (1987). "Exon and Domain Evolution in the Proenzymes of Blood Coagulation and Fibrinolysis." Cold Spring Harbor Symposia on Quantitative Biology: The Evolution of Catalytic Function, LII: 925-932. [40]
- Doolittle, R. F. and Feng, D. F. (1987). "Reconstructing the Evolution of Vertebrate Blood Coagulation from a Consideration of the Amino Acid Sequences of Clotting Proteins." Cold Spring Harbor Symposia on Quantitative Biology: The Evolution of Catalytic Function, LII: 869-874. [41]
- ...all of which goes to show that Cavalier-Smith was not engaging in ad hominem when he wrote of Behe: "What is sad about this book is that the author thinks that he has something new to say and is contributing to science."
Not to pile things on, but upon discovering the foregoing I was inspired to double-check Behe's claims about how often the cilium papers Cavalier-Smith (1978) and Szathmary (1987) were cited in other papers. On page 69, Behe says "The scientific community at large has ignored both contributions; neither paper has been cited by other scientists more than a handful of times in the years since publication.7" Note 7 on page 280 reads, "A search of Science Citation Index shows that each paper receives an average of less than one citation per year."
Well, I don't know if Behe made a mistake or if the Science Citation Index was in 1995 just not adequate on these papers for some reason, but my search of the Science Citation Index Expanded (SCI-EXPANDED)--1945-present on the [http://www.webofscience.com Web of Science], while it lists only six citations of Szathmary (1987), lists 53 citations of Cavalier-Smith (1978). Seven citations are from 1996-present (four of those being in other papers by Cavalier-Smith). For 1995 and before, this leaves 47 citations. Ten of these are in other papers Cavalier-Smith authored or co-authored, leaving 37 citations of Cavalier-Smith (1978) by papers not written by Cavalier-Smith. For those who are counting, that is 2.056 citations per year for the years 1978-1995 inclusive (and including 1978 is being generous to Behe), and while not an overwhelming number, (a) it is higher than "less than one citation per year" and (b) 37 citations (40 to present, and not counting several references in books that I know of) is frankly significantly more attention than the average scientific paper receives.
Out of curiosity, I performed the same search on Cavalier-Smith (1982) and got a result of zero citations. This is clearly a mistake, as not only does Cavalier-Smith reference his 1982 paper in several other papers, but a cursory manual search on Web of Science of the reference lists of other papers reveals that it is cited in (at least) Bermudes et al. (1994), Rizzotti (1995), and Thornhill and Ussery (2000). Obviously there is an error in the Science Citation Index record for the 1982 paper, a good demonstration of the potential risks of relying completely on computer searches.
Cavalier-Smith's 1975, 1987(a), 1987(b), and 1987(c) papers are listed as having 98, 41, 94, and 133 citations respectively, although it appears that self-citation occurs in proportions similar to the 1978 case (self-citation is common in scientific papers).
Finally, Cavalier-Smith recently (November 2001) published in the [http://link.springer.de/link/service/journals/00239/ Journal of Molecular Evolution] a long (40 pages, huge for a journal) comprehensive article on the origin of the first cells, a major update of his 1987(a) paper:
- J Mol Evol 2001 Oct-Nov;53(4-5):555-95
- Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis.
- Cavalier-Smith T.
- Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, United Kingdom.
I attempt to sketch a unified picture of the origin of living organisms in their genetic, bioenergetic, and structural aspects. Only selection at a higher level than for individual selfish genes could power the cooperative macromolecular coevolution required for evolving the genetic code. The protein synthesis machinery is too complex to have evolved before membranes. Therefore a symbiosis of membranes, replicators, and catalysts probably mediated the origin of the code and the transition from a nucleic acid world of independent molecular replicators to a nucleic acid/protein/lipid world of reproducing organisms. Membranes initially functioned as supramolecular structures to which different replicators attached and were selected as a higher-level reproductive unit: the proto-organism. I discuss the roles of stereochemistry, gene divergence, codon capture, and selection in the code's origin. I argue that proteins were primarily structural not enzymatic and that the first biological membranes consisted of amphipathic peptidyl-tRNAs and prebiotic mixed lipids. The peptidyl-tRNAs functioned as genetically-specified lipid analogues with hydrophobic tails (ancestral signal peptides) and hydrophilic polynucleotide heads. Protoribosomes arose from two cooperating RNAs: peptidyl transferase (large subunit) and mRNA-binder (small subunit). Early proteins had a second key role: coupling energy flow to the phosphorylation of gene and peptide precursors, probably by lithophosphorylation by membrane-anchored kinases scavenging geothermal polyphosphate stocks. These key evolutionary steps probably occurred on the outer surface of an 'inside out-cell' or obcell, which evolved an unambiguous hydrophobic code with four prebiotic amino acids and proline, and initiation by isoleucine anticodon CAU; early proteins and nucleozymes were all membrane-attached. To improve replication, translation, and lithophosphorylation, hydrophilic substrate-binding and catalytic domains were later added to signal peptides, yielding a ten-acid doublet code. A primitive proto-ecology of molecular scavenging, parasitism, and predation evolved among obcells. I propose a new theory for the origin of the first cell: fusion of two cup-shaped obcells, or hemicells, to make a protocell with double envelope, internal genome and ribosomes, protocytosol, and periplasm. Only then did water-soluble enzymes, amino acid biosynthesis, and intermediary metabolism evolve in a concentrated autocatalytic internal cytosolic soup, causing 12 new amino acid assignments, termination, and rapid freezing of the 22-acid code. Anticodons were recruited sequentially: GNN, CNN, INN, and *UNN. CO2 fixation, photoreduction, and lipid synthesis probably evolved in the protocell before photophosphorylation. Signal recognition particles, chaperones, compartmented proteases, and peptidoglycan arose prior to the last common ancestor of life, a complex autotrophic, anaerobic green bacterium.
In this paper he repeatedly cites two additional recent papers:
Cavalier-Smith, T (2002a) The neomuran origin of archaebacteria, the negibacterial root of the universal tree, and bacterial mega classification. Int J Syst Evol Microbiol
Cavalier-Smith, T (2002b) The phagotrophic origin of eukaryotes and phylogenetic classifications of Protozoa. Int J Syst Evol Microbiol
...both of which came out in the International Journal of Systematic and Evolutionary Microbiology. Together these three papers constitute some 150 pages, and they serve as only the barest introduction to the topic of the origin of the three domains and their various biochemical systems. For more articles by Cavalier-Smith, click over to PubMed and search on "Cavalier-Smith". Be sure to click on "related articles" for topics you are interested in.
If you are looking for a single scientist to serve as the "Darwin of microbiology" as a counter to Mike "the Paley of microbiology" Behe, it would be Thomas Cavalier-Smith.
Some references on the high variability in the construction of the cilium ("the cilium" isn't IC, just a subset of its components), and on the evolution of the cilium in reference to Behe's arguments
Afzelius, B. A. (1982). �The flagellar apparatus of marine spermatozoa: evolutionary and functional aspects.� Symposia of the Society for Experimental Biology 35(5): 495-519. Link: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=6764047&dopt=Abstract
Cavalier-Smith, T. (1997). �The blind biochemist.� Trends in Ecology and Evolution 12(4): 162-163. (This is Cavalier-Smith's dissection of Behe.)
Miller, K. R. (1999). Finding Darwin's God. New York, Cliff Street Books, pp. 140-143. Link: http://www.amazon.com/exec/obidos/ASIN/0060930497
Stevens, C. (1998). "A Rebuttal of Behe." Produced by Clare Stevens. Accessed online on 3/20/2001. Webpage: http://www.btinternet.com/~clare.stevens/behenot.htm
Some web references on FtsZ-tubulin homology
[http://octem.berkeley.edu/webpage/papers/nature/index.html Tubulin and FtsZ form a distinct family of GTPases] by Nogales et al. (originally in Nature Structural Biology)
[http://www.cellbio.duke.edu/Faculty/~Erickson/FtsZ_tubulin_struct.html Atomic structures of tubulin and FtsZ] by [http://www.cellbio.duke.edu/Faculty/~Erickson/%7ENucleus Harold Erickson]
[http://www2.mrc-lmb.cam.ac.uk/groups/JYL/ftsz.html Structure of the bacterial tubulin homolog FtsZ] by Lowe and Amos
[http://www2.mrc-lmb.cam.ac.uk/groups/JYL/sheets.html Tubulin-like protofilaments in Ca-induced FtsZ sheets] by Lowe and Amos
[http://www.cib.csic.es/~taxol/taxol_en.html Structure & function of homologous proteins tubulin and FtsZ] -- some strong medical applications of this evolutionary deduction
Egelman, E. H. (1998). �Tubulin Family: Kinship of key proteins across phylogenetic domains.� Current Biology 8: R288-R290. Link: http://biomednet.com/elecref/09609822008R0288
Faguy, D. M. and Doolittle, W. F. (1998). �Cytoskeletal proteins: The evolution of cell division.� Current Biology, V8(N10): R338-R341. Link: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9601632&dopt=Abstract
Check out this [http://www.cellbio.duke.edu/Faculty/~Erickson/FtsZ_phylogenetic_tree.html phylogenetictree of FtsZs].
Bacterial[edit]
Description[edit]
The filament is composed of the protein flagellin and is a hollow tube 20 nanometers thick. It is helical, and has a sharp bend just outside the outer membrane called the "hook" which allows the helix to point directly away from the cell. A shaft runs between the hook and the basal structure, passing through protein rings in the cell's membranes that act as bearings. Gram-positive organisms have 2 rings, one in the cell wall and one in the cell membrane. Gram-negative organisms have 4 rings, 2 in the cell wall and 2 in the cell membrane.
The bacterial flagellum is driven by a rotary engine composed of protein, located at the flagellum's anchor point on the inner cell membrane. The engine is powered by proton motive force, i.e., by the flow of protons across the bacterial cell membrane due to a concentration gradient set up by the cell's metabolism (in Vibrio species the motor is a sodium ion pump, rather than a proton pump). The rotor transports protons across the membrane, and is turned in the process. The rotor by itself can operate at 6,000 to 17,000 revolutions per minute (rpm), but with a filament attached usually only reaches 200 to 1000 rpm.
The components of the flagellum are capable of spontaneous assembly in bacterial membranes. Both the basal structure and the filament have a hollow core, through which the component proteins of the flagellum are able to move into their respective positions. The basal structure has many traits in common with some types of secretory pore which have a hollow rod-like "plug" in their centers extending out through the cell wall, and it is thought that bacterial flagella may have evolved from such pores.
Different species of bacteria have different numbers and arrangements of flagella. Monotrichous bacteria have a single flagellum. Lophotrichous bacteria have multiple flagella located at the same spot on the bacteria's surface which act in concert to drive the bacteria in a single direction. Amphitrichous bacteria have a single flagellum each on two opposite ends (only one end's flagellum operates at a time, allowing the bacteria to reverse course rapidly by switching which flagellum is active). Peritrichous bacteria have flagella projecting in all directions.
Some species of bacteria (those of Spirochete body form) have internal flagella that lie between their inner and outer membranes, the rotation of which causes the entire bacterium to corkscrew through its medium.
Anticlockwise rotation of monotrichous polar flagella thrusts the cell forward with the flagellum trailing behind. Periodically the direction of rotation is briefly reversed, causing what is known as a "tumble", and results in reorientation of the cell.
Spirochaetes[edit]
Spirochaetes do all that, but inside their membrane. Instead of spinning the rotor, they spin themselves.
Evolution[edit]
Note[edit]
This section has been almost entirely superceded by:
- Answering the biochemical argument from design, by Kenneth Miller (2003), in: Manson, N. (Ed.), God and design: the teleological argument and modern science, Routledge, London, pp. 292-307.
- The Flagellum Unspun, by Kenneth Miller (2004), in: Dembski, W., and Ruse, M. (Eds.), Debating Design: from Darwin to DNA, Cambridge University Press, New York.
- See also Ken Miller's website. Dembski responded to Miller in an online essay Still Spinning Just Fine on his Design Inference website
- Musgrave, Ian (2004). "Evolution of the Bacterial Flagellum", in: Young, M., and Edis, T. (Eds.), Why Intelligent Design Fails: A Scientific Critique of the Neocreationism, forthcoming from Rutgers University Press, Piscataway, N.J.
- And of course my (Matzke's) review of the evolution of the bacterial flagellum, online at talkdesign, with a background page that should be read first.
However, the below text is still decent background, so I will leave it for the moment.
Introduction[edit]
For the Intelligent Design movement, the bacterial flagellum has now become the equivalent of the eye in 19th century debates. It has become the prime "Icon of ID," despite the fact that Behe only devoted a few pages to it (far less than his discussion of the cilium or other systems). There is no evidence that the bacterial flagellum had any importance at all for helping along the evolution of more complex organisms, for instance; and as discussed below, they represent a "technology" that is actually quite hazardous to eukaryotes.
Evolution of the Bacterial Flagellum[edit]
Evolution of the Bacterial Flagella, a draft FAQ by Ian Musgrave. The fundamental point is that a subset of flagellar components can serve a function as a Type III transport system. Admittedly, all currently known nonflagellar Type III transport systems are for injecting toxins into eukaryotic cells, and are therefore presumably descended from the flagellum, which is likely older than eukaryotes. However, the Type III transport system still proves that the flagellum did not have to come about all at once as Behe argues, as a subset of components has a selectable function. That all known nonflagellar Type III transport systems are disease mechanisms is not shocking as the Type III secretion system was only discovered in 1994 (Cornelis and Gijsegem, 2000) and as our scientific study of eubacteria is significantly biased towards disease-causing organisms for obvious good reasons. We have another rather spectacular case of co-option, where a motility organelle has evolved into a "complex weapon for close combat"
Homologies to Other components[edit]
Two pages in the UCSD transport system database note that the flagellar motor proteins (MotA and MotB) are distant homologs of other bacterial membrane proteins (ExbB-ExbD).
For a few months, the Mot-Exb connection was an isolated mention on a webpage. But in October 2001, an article by Kojima and Blair in Biochemistry made it official. The abstract is here. In their conclusion, the authors write (see the article for a comparative graphic),
The occurrence of significant conformational change in the stator has implications not only for the present-day mechanism but
also for the evolution of the flagellar motor. A membrane complex that undergoes proton-driven conformational changes could perform useful work in contexts other than (and simpler than) the flagellar motor, and ancestral forms of the MotA/MotB complex might have arisen independently of any part of the rotor. We queried the sequence database using the sequence of the best-conserved part of MotA (the segment containing membrane segments 3 and 4) from Aquifex aeolicus, a species whose lineage is deeply branched from other bacteria. In addition to the expected MotA homologues, the search returned a protein sequence from the archaeal species Methanobacterium thermoautotrophicum (protein MTH1022) that shows significant sequence similarity not only to MotA but also to the protein ExbB (Figure 9). ExbB is a cytoplasmic-membrane protein that functions in conjunction with ExbD, TonB, and outer-membrane receptors to drive active transport of certain essential nutrients across the outer membrane of Gram-negative bacteria. The energy for this transport comes from the proton gradient across the inner membrane. Thus, MotA and ExbB are both components of systems that tap the proton gradient to do work some distance away (at either the rotor-stator interface or the outer membrane; Figure 9).
Other features also point to a connection between the Mot and Exb systems. MotA functions in a complex with MotB, which as noted contains the critical residue Asp32 near the cytoplasmic end of its single membrane segment. ExbB functions in a complex with ExbD, which likewise has a single membrane segment with a critical Asp residue near its cytoplasmic end (Asp25 in ExbD of E. coli; ref 59). Although ExbB has only three membrane segments in contrast to the four in MotA, the membrane segments that show sequence similarity have the same topology. The protein TonB is also present in the complex with ExbB and ExbD (59, 60) and would provide an additional membrane segment to round out the topological correspondence (Figure 9). ExbB contains a well-conserved Pro residue (Pro141 in E. coli ExbB) that is the counterpart of Pro173 of MotA. Although MotB and ExbD do not share close sequence similarity apart from the critical Asp residue, in certain positions in the membrane segment the residues most common in MotB proteins are also common in ExbD proteins. Finally, like the MotA/MotB complex the ExbB/ExbD complex contains multiple copies of each protein (61). Together, these facts make a reasonable case for an evolutionary connection between the Mot proteins of the flagellar motor and the Exb proteins of outer-membrane transport (and by extension the TolQ/TolR proteins, which are related to ExbB/ExbD but whose functions are less understood).
...which appears to improve significantly on the situation as stated
in Ian Musgrave's draft flagellum FAQ, "There is no apparent homolog of
the motor (MotAB) in type III secretory systems."
For additional information on homologies see:
The UCSD page on The Type III (Virulence-related) Secretory Pathway (IIISP) Family
In addition, the ATPase involved in flagellar construction is thought to be homologous to this family. Rizzotti, in his (2000) book Early Evolution has cited this in his scenario for the origin of the bacterial flagellum from an F1F0 ATPase. In light of the other homologies mentioned here, this is very unlikely, although there may be a connection between the F1F0 ATPase and transport systems at some very remote level.
Discovery Institute fellow Scott Minnich recently (November 2000) spoke at the DI's "[http://resources.christianity.com/ministries/lifeaudio/main/seriesInfo.jhtml?id=515 Science and Evidence for Design in the Universe]" conference. The talk, "[http://resources.christianity.com/ministries/lifeaudio/main/talkInfo.jhtml?id=1583 Flagella: Tails of Molecular Cooption]", is available in audio format online, and includes the muddled reaction from the audience. Minnich raised the issue of Type III virulence systems, speaking of "malevolent design", and suggested "If we're going to use this [flagellum] as a designed system, we're going to have to use this [Type III virulence system] as well", and "In the original design, they [virulence factors] weren't involved. You can almost look at this type III system as a perversion." Compare to Kathryn Brown's introduction to a Yersinia article in Science, "Why, the people wondered? Was the Plague the work of an angry God? A medieval curse? As it happens, the real culprit was a tiny bacterium: Yersinia pestis." And of course its syringe-like, IC, "designed-looking" virulence system. According to ID, apparently the "angry God" and "bacterium" theory for The Plague aren't as mutually exclusive as one might think.
Some have described the type III virulence systems as a complex weapon designed for close combat (Cornelis and Gijsegem, 2000). So if the IDer did after all design the (eu)bacterial flagellum, we also have the IDer to thank for "front-loading" in the potential for some of the nastiest bacterial virulence systems, including those of Salmonella and Yersinia, the latter being better known as the Bubonic Plague or Black Death. Comparing the explanations given for the virulence of Yersinia between the medievals of the 14th century and the ID theorists of the 21st may be a worthwhile research project.
Some online articles on Type III transport virulence systems and flagella
[http://mmbr.asm.org/cgi/content/full/62/2/379?view=full&pmid=9618447 Type III Protein Secretion Systems in Bacterial Pathogens of Animals and Plants] by Christoph J. Hueck
[http://www.pnas.org/cgi/content/full/95/24/14006 Interactions of Salmonella with host cells: Encounters of the closest kind] by Jorge Galan
[http://micro.annualreviews.org/cgi/content/full/54/1/735 Assembly and Function of Type III Secretory Systems] by Cornelis and Gijsegem
[http://www.cdc.gov/ncidod/EID/vol2no4/mecsas.htm Molecular Mechanisms of Bacterial Virulence: Type III Secretion and Pathogenicity Islands] by Mecsas and Strauss. On the [http://www.cdc.gov Center for Disease Control] webpage.
[http://www.sciencemag.org/cgi/content/full/290/5496/1475 Unlocking the Secrets of the Grim Reaper], by Kathryn Brown, introducing an article on Yersinia in Science.
Archaeal[edit]
Description[edit]
The archaeal flagellum is another prokaryote flagellum that is found exclusively in the archaea (also known as archaeabacteria, depending on whether or not one believes that these prokaryotes constitute a fundamental domain of life (e.g., Woese), or a just a highly-derived bacterium with heavy adaptation to extremophily, particularly thermophily (e.g., Cavalier-Smith)).
The archaeal flagellum is superficially similar to the bacterial (or eubacterial) flagellum; in the 1980s they were thought to be homologous on the basis of gross morphology and behavior (Cavalier-Smith, 1987). Both flagella consist of filaments extending outside of the cell, and rotate to propel the cell.
However, discoveries in the 1990s have revealed numerous detailed differences between the archaeal and bacterial flagella; these include:
- Bacterial flagella are powered by a flow of H+ ions (or occasionally Na+ ions); archaeal flagella are almost certainly powered by ATP.
- Bacterial flagella grow by the addition of flagellin subunits at the tip; archaeal flagella grow by the addition of subunits at the base.
- Bacterial flagella are thicker than archaeal flagella, and the bacterial filament has a large enough hollow "tube" inside that the flagellin subunits can flow up the inside of the filament and get added at the tip; the archaeal flagellum is too thin to allow this.
- Many components of bacterial flagella share sequence similarity to components of the Type III secretion systems (TTSS); the components of bacterial and archaeal flagella share no sequence similarity, however, some components of archaeal flagella share sequence similarity with Type IV secretion systems, also known as Type IV pili.
These differences mean that the bacterial and archaeal flagella are a classic case of biological analogy![]() , rather than homology
, rather than homology![]() ; however, in comparison to the decades of well-publicized study of bacterial flagella (e.g. by Berg), archaeal flagella have only recently begun to get serious scientific attention. Therefore many assume erroneously that there is only one basic kind of prokaryotic flagellum, and that archaeal flagella are homologous to it (an example is Cavalier-Smith (2002), who is aware of the differences in archaeal and bacterial flagellins, but retains the misconception that the basal bodies are homologous).
; however, in comparison to the decades of well-publicized study of bacterial flagella (e.g. by Berg), archaeal flagella have only recently begun to get serious scientific attention. Therefore many assume erroneously that there is only one basic kind of prokaryotic flagellum, and that archaeal flagella are homologous to it (an example is Cavalier-Smith (2002), who is aware of the differences in archaeal and bacterial flagellins, but retains the misconception that the basal bodies are homologous).
Evolution[edit]
The recently elucidated archaeal flagellum is analogous, not homologous, to the bacterial one. In addition to no sequence similarity being detected between the genes of the two systems, the archaeal flagellum appears to grow at the base rather than the tip, and is about 15 nanometers (nm) in diameter rather than 20. Arguing for the design of the eubacterial flagellum, Minnich and other ID advocates place much weight on the lack simpler homologs that are known to be non-derived. Interestingly, sequence comparison indicates that the archaeal flagellum is homologous to Type IV pili (pili are nonmotile filamentous structures outside the cell) and better yet, to twitching motility systems, which allow the cell to crawl along a surface. These systems are in turn homologous to the Type II transport system, which is the conclusion of the general secretory pathway.
For references and more info on the archaeal flagellum, see almost anything that Ken Jarrell has written, e.g. the articles linked from [http://info.queensu.ca/micr/faculty/jarrell/jarrell_laboratory.htm The Jarrell Laboratory] hompage.
Also see this online version of "A twisted tale: the origin and evolution of motility and chemotaxis in prokaryotes", by Faguy and Jarrell
"Recent excitement about the Archaea", by Jarrell, in Bioscience
Finally, in a 1998 article by Bayley and Jarrell, they write,
We feel that the discoveries of archaeal flagella-related putative gene products with similarity to type IV pilus accessory proteins indicate that the archaeal flagella also share this common origin and have evolved it to function as the primary motility apparatus. Although the function of the common origin can only be speculated, this system must predate the last common ancestor of extant life. ("Further Evidence to Suggest That Archaeal Flagella Are Related to Bacterial Type IV Pili", Journal of Molecular Evolution, 46: 370-373, 1998)
Summary[edit]
In conclusion, there is no reason to think that the natural evolution of the various flagella is impossible or wildly unlikely, and therefore it is not imperative to think that they were designed by intelligent intervention. Testable outlines exist for the origin of each of the three motility systems, and avenues for further research are clear:
- For prokaryotes, these avenues include the study of secretion systems in free-living, nonvirulent prokaryotes.
- In eukaryotes, the mechanisms of both mitosis and cilial construction, including the key role of the centriole, need to be much better understood. A detailed survey of the various nonmotile appendages found in eukaryotes is also necessary.